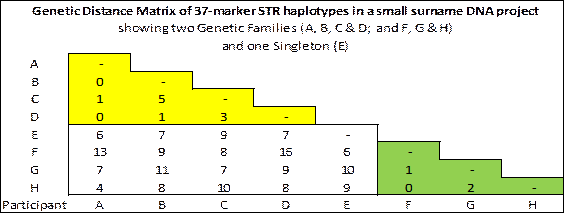Matching and grouping in surname DNA projects
From ISOGG Wiki
The matching and grouping of Y chromosome DNA test results are important responsibilities of administrators of Surname DNA projects. Administrators need an understanding of both responsibilities, regardless of how they may choose to display their projects' Y-DNA Results (choices which are discussed here). Alas both are challenges that are complicated by the lack of standardised terminology and, when handled responsibly, by the need for a degree of judgement rather than being treated as purely mechanical exercises. They are thus not easily explained, and personal preferences of individual administrators may legitimately differ.
Contents
Terminology
For the purpose of this article the following interpretations are used:
- Matching: the process of comparing two Y-STR haplotypes (aka genetic signatures) to determine if they are likely to share a common patrilineal ancestor within the genealogical timeframe.
- In surname DNA projects a match is considered to exist when a comparison of the STR haplotypes of two testees suggests there is a high probability of them sharing a common patrilineal ancestor within the genealogical timeframe.
- A "match" includes, but is not limited to, an exact match, a close or near match, and a match as defined in Family Tree DNA's Matches pages; where the latter is discussed it is prefixed by "FTDNA". Individual administrators legitimately define match in different ways.
- Grouping: the process of arranging/categorizing/classifying two or more participants in a project who share some common feature. Such groupings may be known as:
- Group or Cluster: any grouping of participants sharing some common feature;
- Genetic family: a grouping of STR haplotypes thought to share a common patrilineal ancestor within the genealogical timeframe, immaterial of genealogies or surname.
- A singleton is a haplotype which does not match any other haplotype in a surname project.
NB
1. By "genealogical timeframe" (aka "surname era") is meant the timespan when the surname concerned has been hereditary. In Ireland hereditary surnames were uncommon before the 10th century, in England before the 11th century, and in Scotland before the 13th century. Many surnames have existed for much shorter periods.
2. Genetic families generally comprise two or more of a surname project's STR haplotypes. Some administrators require three haplotypes, for greater stringency; some administrators allow groupings of one if the haplotype is accompanied by noteworthy genealogical data.
3. Terms such as "Lineage" and "Patriarchal line" are sometimes used synonymously with "genetic family". The application of such terms from conventional genealogy to genetic genealogy can cause confusion; in particular participants with matching haplotypes (see below) may not descend from the patriarch identified by conventional genealogy but from a different lineage that shares an older common ancestor.
4. The interpretations above are not universally accepted.
Matching
Several tools may be used (e.g. FTDNA's "Y-DNA Matches" pages, Genetic Distance, FTDNA's TiP tool, TiP Score), each quite legitimately, to determine whether or not two STR haplotypes are matches as interpreted above, i.e. if two participants share a common patrilineal ancestor.
FTDNA's "Y-DNA Matches" pages
On the personal page of each testee on their website Family Tree DNA list the Y-STR Matches of that testee with the other testees in the various projects in which the testee participates, or with the other testees in FTDNA's “Entire Database”. Matching can be selected at resolutions of 12, 25, 37, 67 or 111 markers (if the common test resolution permits).
FTDNA deem another testee to be a "match" if the two haplotypes are:
- Genetic Distance of 1 or less at 12 markers (within project, otherwise 0);
- Genetic Distance of 2 or less at 25 markers;
- Genetic Distance of 4 or less at 37 markers;
- Genetic Distance of 7 or less at 67 markers;
- Genetic Distance of 10 or less at 111 markers.
NB.
1. By "Genetic Distance" FTDNA mean a hybrid genetic distance as adopted on 20 Dec. 2012.
2. Only those testees who have signed the FTDNA Release Form will appear in these Matches pages.
It is important to remember that:
1. When FTDNA's lists indicate that two haplotypes "match", this is not a guarantee they share a common patrilineal ancestor: whichever matching tool is used, the result is always a matter of probability - see [FAQ:id919].
2. FTDNA’s matching criteria are irrespective of surname, and thus often include, especially if the "Entire database" option is selected at a resolution of 12 or 25 markers, many matches of STR haplotypes of testees with dissimilar surnames (see section below – "Matching of STR Haplotypes of Testees with Dissimilar Surnames").
3. Even for comparisons of two haplotypes of testees with the same or similar surnames, some experienced project administrators consider that the criteria used in FTDNA’s Matches pages are too lax, while others consider they are too stringent.
FTDNA's Matches pages are thus very handy to use, but are rather coarse and simplistic and can exclude haplotypes of participants who share a common ancestor but have had an unusually high number of mutations.
Genetic distance determined by the project administrator
The criteria used in FTDNA's Matches pages may also be written as 11/12, 23/25, 33/37, 60/67 and 101/111, or 1:12, 2:25, 4:37, 7:67 or 10:111, or may be called the "1, 2, 4, 7, 10" "rule of thumb".
Some project administrators prefer a more stringent rule of thumb for matching two testees with similar surnames, for example "-, 0, 3, 5", where 12-marker tests are deemed inadequate and genetic distances of 0, 3 or less, and 5 or less are required for a match when the highest resolution shared by the two haplotypes is 25, 37 or 67 markers respectively.
Conversely other project administrators accept a less stringent rule of thumb, for example "1, 3, 5" at 12, 25 and 37 markers respectively.
But as matching is a matter of probability no particular rule of thumb is "right" or "wrong".
NB
1. Most administrators are implicitly and often unconsciously using the current FTDNA hybrid genetic distance; many have probably retained the former FTDNA hybrid genetic distance, unaware that FTDNA changed their definition on 20 December 2012; some may prefer stepwise or infinite-alleles genetic distances. In practice, however, the differences between the various definitions of genetic distance are only of limited importance.
2. Whatever "rule of thumb" and limiting criteria an administrator may choose to adopt, all matches based on genetic distance criteria are an approximation as they assume that all markers have the same average mutation rates.
3. These rules of thumb apply when matching haplotypes of participants with the same or similar surnames. Most surname project administrators adopt a more stringent rule of thumb for comparing dissimilar surnames. (See the section on "Matching of STR haplotypes of testees with dissimilar surnames below.)
FTDNA TiP tool
TiP, FTDNA's Time Predictor tool, is FTDNA's most advanced matching tool. It incorporates their best estimates of the average mutation rates for individual markers and gives the probability of two haplotypes matching over a given number of generations. These probabilities can be modified to take account of paper trail data.
The use of TiPs has not proved popular with novice administrators because of their complexity, or with some more experienced administrators because FTDNA have not disclosed details of their derivation.
TiP Score
This is a simplified adaptation of TiP that has been adopted by a few administrators. TiP Score is defined as the 24-generation, no-paper-trail TiP, at the highest common resolution of the two haplotypes being compared. It is an arbitrary selection of one of the many TiPs that can be derived when two haplotypes are compared, and thus sacrifices much of the potential of the TiP concept. On the other hand it is a very simple and robust yardstick that administrators can conveniently use when establishing a genetic family or adding additional haplotypes to an existing genetic family, by referring to https://gap.familytreedna.com/genetic-distance-report.aspx or https://gap.familytreedna.com/tip-report.aspx.
The project administrator may adopt a TiP Score threshold of 60% or 80% to determine whether or not two haplotypes with the same or similar surnames constitute a match. For haplotypes with dissimilar surnames see the section on "Matching of STR haplotypes of testees with dissimilar surnames" below.
TiP Scores can be further simplified by dispensing with the decimal points – as they are only relative indicators, probabilities to two decimal places give a misleading impression of accuracy. Unlike genetic distances, TiP Scores make clear the probablistic nature of matching, and they can be used for matching haplotypes with differing resolutions. Further benefits of TiP Scores will become apparent under "Grouping" in section 3 below.
Matching of STR haplotypes of testees with dissimilar surnames
The matching criteria considered above are only valid when seeking to determine whether two testees with the same or similar surnames share a common patrilineal ancestor within the genealogical timeframe. FTDNA give detailed advice on probabilities of matches of two testees sharing a surname, but give no explicit guidance on "matching" two testees with dissimilar surnames. For the latter the same principles apply, except that most administrators consider it necessary to adopt some more stringent criteria.
One reason for dissimilar surnames appearing on FTDNA's "Matches" pages are NPEs and the non-hereditary use of surnames, which do share a common patrilineal ancestor. To allow for these possibilities some project administrators require such matches to be 35/37 or 63/67 or better, i.e. they apply a "-, -, 2, 4" rule of thumb, or a TiP Score of 90% or 95%, together with some evidence of an NPE having occurred, for example from genealogical evidence of a confirmed or suspected NPE, or evidence that the families with the two surnames were neighbours at some period.
Another reason for dissimilar surnames appearing on FTDNA's "Matches" pages is convergence, the apparent matching of STR haplotypes due to random mutations of individual markers, which do not share a common patrilineal ancestor within a genealogical timeframe. To reduce the risk of misleading matches caused by convergence the World Families Network website recommends disregarding all FTDNA's listed matches of haplotypes of participants with dissimilar surnames below the resolution of 37 markers, while some project administrators only consider such matches at a resolution of 67 markers or more. SNP tests are increasingly being used to show that even though two testees may share, for example, a 35/37 or 63/67 match, if they belong to different sub-clades they cannot share a common patrilineal ancestor within the genealogical timeframe.
NB
1. Such a negative SNP test may no longer apply when comprehensive Y-chromosome sequencing tests using next generation sequencing become the norm.
2. The fact that two testees do share a common SNP does not necessarily mean they share a common paternal ancestor within the genealogical timeframe.
Alas at present there is no reliable means of determining whether dissimilar surnames appearing on FTDNA's "Matches" pagss are due to NPEs, non-hereditary surnames or convergence.
Grouping
The grouping of participants' Y-DNA results in a surname project is one of the most important responsibilities of the project administrator. However, as with "matching", there is no right or wrong way of doing this: the optimal choice will depend on the knowledge of the administrator, the size of the project, the diversity of the haplotypes tested, the particular haplogroups involved, and the availability of relevant genealogical background. Choices include:
By genealogical feature
Grouping by some genealogical feature such as the spelling of the surname of the participant or his earliest ancestor or by the place of residence of the participant or his earliest ancestor may be suggested by the goals of the project and initially may appear tidy and logical, but each have limitations: the spelling of nearly all surnames has been inconsistent, especially for migrants and before the mid 19th century (a common finding of surname DNA projects is that the modern spelling of a surname is an unreliable guide to its origins); genealogical and geographical records are often limited to a few centuries, and/or may be inaccurate. More importantly, all such criteria fail to address the benefits of undergoing DNA tests, and may actually disguise the value of the results.
NB. As the main objective of most surname projects is to match genetic and conventional "paper trail" genealogy, it may appear counterintuitive not to use relevant genealogical data to assist in the grouping of haplotypes. However experience with large projects has shown it is safest to initially base grouping solely on genetic evidence. But once haplotypes have been grouped then it is quite appropriate to name/label groupings after paper trail characteristics of some of its members (see the section on the "Naming and labelling of groupings" below).
By haplogroup
Grouping the project's haplotypes, e.g. A, E, F, R1b etc., as tested for or predicted from STR results by FTDNA. This choice was popular when only 12-marker tests were available, and even today this criterion remains the simplest choice and the easiest to administer.
By subclade
Grouping by subclade is a more refined version of grouping by haplogroup which involves breaking down the haplogroups into more precise groupings. This is particularly desirable when haplogroup groups are large, e.g. haplogroup R1b. Grouping can be carried down to the level of FTDNA's predicted haplogroups, e.g. R1a1b2 or R-M269.
NB. Labelling by subclade may be carried down to the level of terminal SNPs after grouping by genetic family (see below).
The above groupings are quite legitimate, but all fail to capitalise on the potentials offered by the higher resolution Y-STR test results that are now available.
By genetic family
This recommended choice is the grouping of the project’s haplotypes. Conceptually this is straightforward – when a project administrator deems by their chosen matching criteria that two STR haplotypes are a match, then they form a genetic family and as such are considered to share a common ancestor within the genealogical timeframe. A third and subsequent matching participants are added similarly.
Alas problems can arise when a potential new participant is considered but the matches are conflicting, for example when participant "A" is a match with both "B" and "C", but "B" and "C" do not match each other. Several tools may be used to help clarify such dilemmas:
Genetic Distance Matrix. All the genetic distances between the potential members of a genetic family are entered in a matrix, such as in the example below, and the project administrator makes a judgement as to how to define the "boundaries" of the genetic family. For example an administrator using the 4/37 rule of thumb might choose to relax it for the matching of haplotypes B & C and A & H in the matrix below:
The above matrix was created manually.
For large genetic families Dean McGee's Y-DNA Utility can be used to create a genetic distance matrix automatically. Some administrators find this utility difficult to use, but perseverance can be rewarding - see for example http://blairdna.com/group01.html.
Each genetic distance matrix can only be used as an aid for grouping haplotypes of the same resolution. An additional matrix is needed, for example, for grouping 67-marker haplotypes.
Modal haplotype. The project administrator determines the modal (most commonly occurring) value of each marker of the members of the genetic family by inspection, and this modal haplotype is used for comparisons with marginal members for the genetic family. This tool avoids the need for constructing and using a genetic distance matrix, and is justified on the basis that the modal haplotype is the nearest available approximation to the haplotype of the common ancestor from whom all members of the genetic family are descended.
NB. Because of sampling bias,genetic drift and/or founder effect the modal haplotype may not be the same as the haplotype of the common ancestor.
Modal participant. Like the modal haplotype above, except that instead of identifying the theoretical modal haplotype of the genetic family, the member of the genetic family whose haplotype is nearest the modal value is chosen instead. This is an approximation that is easier to establish and is necessary when using TiPs and TiP Scores. To be most effective the modal participant is placed as the first line of the genetic family, and other members of the genetic family can be listed in order of TiP probability, even when they have different resolutions. However these refinements are not possible for test results presented on the FTDNA public pages.
NB. As the genetic family grows in size, the modal haplotype and the modal participant will converge.
Terminal SNPs. By definition two participants in the same genetic family cannot have conflicting terminal SNPs. It is thus desirable to use a SNP test to resolve a marginal grouping.
Cladograms. In theory cladograms can also be used, but for most project administrators these tools are too complex for basic grouping of Y-STR test results.
All the above tools are iterative, and their application needs reviewing from time to time as the project grows in size. Some latitude and flexibility may be appropriate. In practice the number of marginal matches grows at a slower rate than the number of well matched participants.
Sub-division of genetic families. Except when FTDNA public pages are used, administrators may attempt to sub-divide some of their genetic families. These sub-divisions will all share a common ancestor, but within each sub-division there is closer matching, typically of individual markers, of participants who share a more recent common ancestor. Tools for determining such sub-groups include unusual markers, cladograms, etc.
Ungrouped haplotypes (aka unassigned, unmatched or waiting haplotypes)
Whatever grouping choice is made there will inevitably be some residual haplotypes to be listed separately. This may arise because:
(a) the haplotype does not match any other haplotype in the project;
(b) too few markers have been tested to make a reliable grouping; and/or
(c) the administrator has not yet allocated newly joined participants to a particular group.
The former are sometimes known as singletons. Singletons are often "embryo" genetic families, awaiting a new participant with a similar haplotype to join the project, but some may be non-paternity events (NPEs), and some may even be the last surviving member of a surname lineage. Like marginal matches, the number of singletons grows at a slower rate than the number of matched participants.
Unless results are displayed FTDNA public pages, (a) and (b) above may be subdivided by haplogroup.
Excluded participants
Administrators of surname projects may elect to exclude certain categories of participant from the presentation of their project test results:
(a) Participants with haplotypes of 12, or even 25, markers or less.
(b) NPEs, i.e. participants who do not bear the project surname even though their haplotype does match that of one or more participants who use the project surname, even with the more stringent criteria suggested above. Alternatively (as happens in the majority of projects), such participants may be included in the main groupings as relevant, but with some distinctive font.
(c) Participants who joined the project in error or who are visiting the project at the discretion of the project administrator, i.e. participants who do not bear the project surname and whose haplotype does not match that of any participant who uses the project surname.
(d) Participants who have only autosomal DNA or mitochondrial DNA test results. Many surname projects disregard such test results as they have little or no relevance to surname evolution, origins, etc.
NB. Exclusion from "open" surname projects may be achieved by the administrator excluding the Y-DNA results from display or by asking such participants to leave the project. Exclusion from "closed" surname projects may be achieved by the administrator denying them participation in the project.
Naming and labelling of groupings
All the above groupings may be named, or labelled, as the project administrator may prefer, for example by some alphabetical or numerical code (e.g. in the sequence the groupings were first identified), by some compound code such as R1b-01, R1b-02, R1b-03 etc., or by geographical origin. However, several considerations are important:
- Ideally all naming/labelling should be as self-explanatory as possible, making full use of the "Description" available on FTDNA's public pages, for example including the geographical origin of the genetic family, if known.
- If results are to be displayed using the FTDNA public pages (see Displaying Y-DNA Results), then the naming/labelling chosen should be compatible with the alphanumeric sequence into which the names/labels will be automatically sorted (e.g."A" will appear below "1", "A2" will appear below "A11", and "Welsh branch" will appear below "Ungrouped" etc.)
- If genealogical lineage or patriarchal data are included in the description, then it should be realised that the DNA project may reveal lineages additional to that discovered by conventional research. It may thus be astute to use labels that distinguish between genetic groupings and genealogical lineages; for example if a genealogical lineage is known by the town where the lineage resided, the genetic family might be better known by the region where they resided.
- If grouping is by genetic family and labelling is by subclade down to terminal SNP level, then after the terminal SNP of one member of the genetic family has been determined it will not be necessary to undertake SNP tests for other members of the genetic family unless a particular participant's membership of the genetic family is in doubt.
Hopefully the new comprehensive Y-chromosome sequencing tests will help with the sub-division of genetic families.
Conclusion
The novice project administrator reading this page may well feel daunted by the complexities and uncertainties discussed but experience has shown that these issues will recede as their project grows in size and self-confidence is acquired.
Further reading
- How to group surname project members by Maurice Gleeson, DNA and Family Tree Research, 28 January 20201.
- Grouping participants by John Blair.
- Building a mutation history tree with STR data by Maurice Gleeson, The Gleason/Gleeson DNA Project blog, 13 August 2015.
- Genetic distance, genetic families, and mutation history trees by Maurice Gleeson, The Gleason/Gleeson DNA Project blog, 7 August 2015.
- Criteria for allocating members to specific genetic families by Maurice Gleeson, Farrell DNA Project blog, 20 January 2015.
- Y-DNA project grouping with genetic distance. Dave Hamm. Hamm Country Blog, 16 April 2009.
- FTDNA Genetic Distance & Family Group Assignments by Georgia Kinney Bopp, 2 August 2005.