Pangenome
From ISOGG Wiki
A pangenome (or pan-genome or supragenome) attempts to describe all genes and genetic variations found within a given species or subspecies. First conceived in 2005 with analyses in bacteriology, it intersects the fields of biology, computer science, and applied mathematics.[1][2][3]
“In simple terms, the pangenome concept is the realization that the genetic repertoire of a biological species, i.e., the pool of genetic material present across the organisms of the species, always exceeds each of the individual genomes and can be, in several cases, "unbounded": an open pangenome.
“This notion was conceived in 2005 as an unexpected, data-driven outcome of the comparative analyses of a few bacterial genomes. This early example of big data in biology—in which a mathematical model, developed to address a practical question in vaccinology, transformed established concepts—opened biology to the unbounded.”
—Hervé Tettelin and Duccio Medini, The Pangenome[3]
The current reference genome
The current human reference genome, GRCh38, the initial draft of which was published in 2001, is a mosaic of multiple people, but roughly 70% of this singular reference genome was contributed by a male from Buffalo, New York, sample "RP11," whose genome was sequenced after he responded to an advertisement for volunteers in The Buffalo Evening News.[4][5][6][7] The remaining 30% of the GRCh38 reference genome is 23% drawn from 10 other individual samples, and 7% from over 50 additional sources. In GRCh37—published in 2009 and superseded by GRCh38 in December 2013, though for genetic genealogy most testing and reporting companies still use GRCh37—72% of the genome came from sample RP11; 23% from 10 other samples, and 5% from over 50 additional sources.[5][8]
Over the course of multiple major versions and minor release iterations, the human reference genome has refined data and made corrections to that reference assembly, but it has remained essentially a flat set of data extrapolated from a small cohort of individuals of primarily European descent. Described a different way in 2019 by Sara Ballouz, Alexander Dobin, and Jesse A. Gillis, the current reference genome is not a baseline, but rather more accurately would be categorized as a type specimen.[5] Additionally, because conventional short-read DNA sequencing made it impossible to annotate approximately 6% of the human genome (roughly 185 million base pairs), until newer, hybridized sequencing methods could be brought to bear in 2021 and 2022, a significant portion of the genome remained obscured.[9]
First full sequencing published in 2022
The first full sequencing of a human genome was formally published in 2022 by the Telomere-to-Telomere Consortium, but it likewise represents only a single individual.[9] Some genetic variants occur more often in some populations than others, so by using a single reference genome a bias is created in calling that particular sequence a "reference" even though that may not be true for different, major global populations.[7] Many members of the Telomere-to-Telomere Consortium were simultaneously working as part of the Human Pangenome Reference Consortium—including Heng Li, Karen Miga, Adam Phillipy, Winston Timp, and others—to leverage the improved long-read and nanopore sequencing techniques in moving toward a human pangenome reference model. [10]
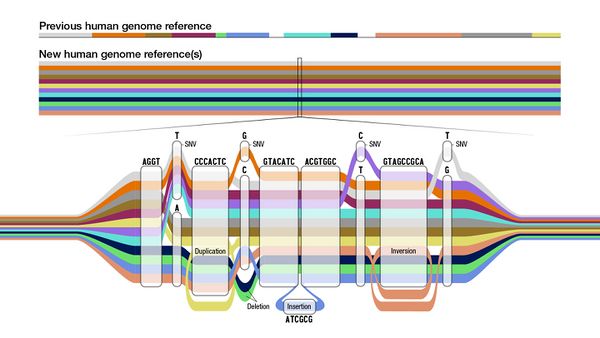
The initial human pangenome reference
The first draft of a human pangenome reference was formally published in Nature on 10 May 2023.[10] This initial release used "47 phased, diploid assemblies from a cohort of genetically diverse individuals" and is intended to be expanded upon in a planned Human Pangenome Reference Consortium panel capturing a better picture of global diversity from 700 haplotypes of 350 sequenced individuals...a massive increase in breadth from the small cohort size our single-reference model has worked with.[10] The pangenome draft adds "119 million base pairs of euchromatic polymorphic sequences and 1,115 gene duplications relative to the existing reference GRCh38. Roughly 90 million of the additional base pairs are derived from structural variation. Using our draft pangenome to analyse short-read data reduced small variant discovery errors by 34% and increased the number of structural variants detected per haplotype by 104% compared with GRCh38-based workflows..." It further adds "3.7 million additional single-nucleotide polymorphisms (SNPs) in regions non-syntenic to GRCh38."[10]
The NIH's Genome Reference Consortium had anticipated releasing GRCh39 by 2022, but put that on indefinite hiatus while work by the Telemore-to-Telomere Consortium and the Human Pangenome Reference Consortium continued. Posted on their website, the organization states that, "[We] have decided to indefinitely postpone our next coordinate-changing update (GRCh39) while we evaluate new models and sequence content from ongoing efforts to better represent the genetic diversity of the human pangenome, including those of the Telemore-to-Telomere Consortium and the Human Pangenome Reference Consortium."[12]
Implications of pangenomics for genetic genealogy
Given that that the direct-to-consumer testing market for autosomal DNA uses inexpensive microarray testing and is still rooted in the GRCh37 reference assembly that was deprecated almost a decade ago, it is unlikely that emergent sequencing technologies and the move toward a mature human pangenome reference will have appreciable impact on genetic genealogy in the near term. However, the underlying reference assembly is foundational to much of what we do with DNA for genealogy, from identifying segment start and end positions to the basis upon which centiMorgan estimations are calculated. The more inclusive, better refined, and more accurate the genome reference model, the greater precision it can provide us in comparing and evaluating our DNA matches.
References
- ↑ Wikipedia contributors, "Pan-genome," Wikipedia, The Free Encyclopedia, en.wikipedia.org/wiki/Pan-genome (accessed 12 May 2023).
- ↑ Tettelin, Hervé, Vega Masignani, Michael J. Cieslewicz, Claudio Donati, Duccio Medini, Naomi L. Ward, Samuel V. Angiuoli, et al. "Genome Analysis of Multiple Pathogenic Isolates of Streptococcus Agalactiae: Implications for the Microbial 'Pan-Genome.'" Proceedings of the National Academy of Sciences 102, no. 39 (19 September 2005): 13950–55. DOI: 10.1073/pnas.0506758102; Open Access (accessed 12 May 2023).
- ↑ 3.0 3.1 Tettelin, Hervé and Duccio Medini, editors. The Pangenome: Diversity, Dynamics and Evolution of Genomes (Cham, Switzerland: Springer Nature Switzerland AG, 2020). ISBN 978-3-030-38280-3; eBook: ISBN 978-3-030-38281-0. Open Access online: link.springer.com/book/10.1007/978-3-030-38281-0 (accessed 12 May 2023).
- ↑ Lander, Eric S., Lauren M. Linton, Bruce Birren, Chad Nusbaum, Michael C. Zody, Jennifer Baldwin, Keri Devon, et al. "Initial Sequencing and Analysis of the Human Genome." Nature 409, no. 6822 (February 2001): 860–921. DOI: 10.1038/35057062; Open Access (accessed 12 May 2023).
- ↑ 5.0 5.1 5.2 Ballouz, Sara, Alexander Dobin, and Jesse A. Gillis. "Is It Time to Change the Reference Genome?" Genome Biology 20, no. 1 (9 August 2019): 159. DOI: 10.1186/s13059-019-1774-4; Open Access (accessed 13 May 2023).
- ↑ Massive Science (massivesci.com) and NIH/NHGRI (genome.gov). The Human Pangenome, video produced for and supported by the National Human Genome Research Institute (Vimeo: 2020). Open Acess: [vimeo.com/massivesci/pangenome https://vimeo.com/massivesci/pangenome], 5 minutes 30 seconds (accessed 13 May 2023).
- ↑ 7.0 7.1 Howe, Nick Petrić, and Shamini Bundell, hosts. "Nature Podcast." Nature (online; 10 May 2023). DOI: 10.1038/d41586-023-01579-9 (accessed 13 May 2023).
- ↑ Genome Reference Consortium. "How many individuals were sequenced for the human reference genome assembly?" Frequently Asked Questions, online: [https://www.ncbi.nlm.nih.gov/grc/help/faq/#human-reference-genome-individuals www.ncbi.nlm.nih.gov/grc/help/faq/ (accessed 13 May 2023).
- ↑ 9.0 9.1 Nurk, Sergey, Sergey Koren, Arang Rhie, Mikko Rautiainen, Andrey V. Bzikadze, Alla Mikheenko, Mitchell R. Vollger, et al. "The Complete Sequence of a Human Genome." Science 376, no. 6588 (April 2022): 44–53. DOI: 10.1126/science.abj6987; Open Access (accessed 12 May 2023).
- ↑ 10.0 10.1 10.2 10.3 Liao, Wen-Wei, Mobin Asri, Jana Ebler, Daniel Doerr, Marina Haukness, Glenn Hickey, Shuangjia Lu, et al. "A Draft Human Pangenome Reference." Nature 617, no. 7960 (May 2023): 312–24. DOI: [https://doi.org/10.1038/s41586-023-05896-x 10.1038/s41586-023-05896-x; Open Access (accessed 10 May 2023).
- ↑ National Institutes of Health (NIH). "Scientists Release a New Human 'Pangenome' Reference," News Release 10 May 2023. [www.nih.gov/news-events/news-releases/scientists-release-new-human-pangenome-reference https://www.nih.gov/news-events/news-releases/scientists-release-new-human-pangenome-reference] (accessed 12 May 2023).
- ↑ The National Center for Biotechnology Information, the National Institutes for Health. The Genome Reference Consortium website: www.ncbi.nlm.nih.gov/grc (accessed 12 May 2023).
