Full Genomes Corporation
From ISOGG Wiki
|
| |
| Type | Inc. |
|---|---|
| Industry | Genetic genealogy |
| Founded | 2013 |
| Founder(s) | Justin Loe, Leon Kull, Abdullah Marafi |
| Headquarters | Rockville, Maryland, USA |
| Area served | International |
| Products | Genome NGS Sequencing, Interpretation (SNPs, STRs, Phylogeny) |
| Website | www.fullgenomes.com |
Full Genomes Corporation was founded in 2013 to make next generation sequencing technology available for the DTC (direct to consumer) market focusing on the Y chromosome. The founders are well known citizen scientists and active in the genetic genealogy community. The headquarters are in Rockville, Maryland, in the USA.
Contents
Team
- Justin Loe, MS, CEO [1]
- Leon Kull, Director Data Analysis
- Ian Burbulis, Ph.D., Scientific Advisor and Director
Two different teams are available to do quality control and two different groups can handle kit shipping.
Y-DNA comprehensive sequencing
As of August 2017 2,700 samples have been sequenced or analyzed (vs 1,800 in May 2016).[2] By November 2017 71,000 SNPs had been discovered.[3]
Y Elite 2
Introduced in May 2015. FGC designs, since 2016: 2.1c (since June 2016), 2.1b (batch 8001-8003,8006), 2.1a (batch 9008,8004,8005); Y-Elite 2.0 (FGC design, 2015-2016); Y Elite 2.1 (September 2017 onwards?)
- $425
- length coverage: 14 megabases
- read length: 150 bp, coverage ~80x
- supplier: Omega Bioservices and another, ~99% quality control success
- replaces: Y Elite 1 (100 bp BGI) and Y Prime (100 bp)
- features: better SNP calling and better STR calling quality
Product history
Y Elite 1.0 (2013-2015, BGI design): FGC introduced this service DTC (Direct To Consumer) in spring 2013. The test is known as the Y Elite. Key information (see also Y-DNA SNP testing chart):
- $850 USD (introduced at $1,499, for early birds $1,299, later lowered to $1,250, $1,099, $999, $975, $850)
- Sequencing is provided by the BGI in Hong Kong.
- Raw data (BAM file): Read length 100 base pairs, coverage 50x, approximately 20-25 million bp are called from the ca. 58 million Y-chromosome bp (the centromere and the huge q12 region are not included), ca. 14 million bp with reliable mappings (high quality SNPs)[4][5]
- SNP Report: approx. 1700 "usable" SNPs reported, check for known Y-SNPs, Y-DNA haplogroup assignment, Report of private (new) SNPs, in common haplogroups 25-40 private SNPs are found
- YFull evaluation: confidence regions length averaged at 23 Mbp. With average mutation rate 0.82 ∙ 10^-9 per year per bp the resolution is 53 years per mutation (for comparison BigY has 10.31 Mbp and 118 years per mutation). [6]
- Y-STR Report: 300+ Y chromosome STRs (Markers)
- Approx. 99% coverage of the mtDNA sequence
Y Prime (2014-2015, FGC design): was introduced in July 2014. Costs were $750 (originally priced at $625, $589 with an introductory price at launch of $599. Subsequently raised to $650).
- Raw data (BAM file): Read length 100 base pairs
- See: Full Genomes launches Y Prime - a new Y chromosome sequencing product by Debbie Kennett. Cruwys News blog, 26 July 2014.
BAM-file analysis
FGC for $50 provides an analysis of third party BAM files and provides the same interpretation results (SNP, STR) as mentioned above. For details see:
- A new BAM file analysis service from Full Genomes Corporation and a special offer on the FGC test by Debbie Kennett, Cruwys News blog, 26 April 2014.
Whole genome tests
100x and more: Extra-high coverage
Introduced in December 2015
- $5350 USD (introduced for $4750 USD).
20x and more: High-coverage
30x
Introduced in summer 2014.
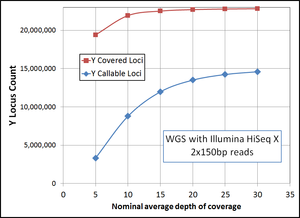
- read depth averages around 20-30. Coverage seems to be well over 90% of the sequenceable genome, probably close to 95%.[7]
- $1425 USD (introduced at $1800 for early birds, later $1850, $1650)
Upgrade costs: $55 per 1x above 30x, 100x: $5350 - Y chromosome data (YFull): Length coverage: 22,858,252 bp, Mean depth coverage:21.38X, Median depth coverage:12X [8]
- Based on dbSNP 142 expected to cover over 100 million known variant sites (mostly SNPs) plus novel variants identified in the ~3 billion nucleotide sites in the human genome; ca. 4.7 million sites called with variation from the reference sequence. [9]
- Data will be delivered by hard drive (100 gigabytes or more of raw data)
Long Read Chromium WGS
$2950. Introduced in September 2016.
7x to 19x: Medium-coverage
- 15x coverage $795 introduced in January 2016.
- 10x coverage $725 (introduced for $675, later $745). Upgrade: 10x -> 15x is $170, 10x -> 30x is $925. Introduced in July 2015. No beta results yet available.
150 bp , ~20+ mbp Y
up to 6x: Low-coverage
Introduced in July 2015. No beta results yet available.
- 4x coverage $395 (introduced for $325, later $350, $375). Upgrade: 4x -> 10x is $330. Use order code: WGS-4xcoverage. Expected similar to 1000 Genomes (4x and higher).
- 2x coverage $280 (introduced for $225, later $250). Upgrade: 2x -> 4x is $115. Use order code: WGS-2xcoverage
Exome sequencing
Exome sequencing was introduced at the end of September 2015. Two tests are available:
- 33x for $775
- 120x for $995
Single cell sequencing
Introduced in spring 2016. Expected primary customers are academic centers who target body regions and cell genome variance for studies. [10] Sequencing of old DNA samples or decayed samples might also be use cases (picograms possible, ie 1 cell of DNA is enough).
Sample collection
Normal procedure: saliva samples (Biomatrica DNAgard). On request further methods [11]:
- blood samples (with lab for QC)
- cheek scrub brush samples
- samples from third party companies
Reviews
- Full Genomes WGS reviews from the DNAtesting choice website
- La génétique - le cas FULL GENOMES - ADN Y (in French) by Joss Ar Gall, Le Gall de Basse Bretagne et d'ailleurs..., 29 March 2014.
- First Look at the Full Genomes Y-Sequencing Results from Itaï Perez. Guest post on CeCe Moore's Your Genetic Genealogist blog, 3 November 2013.
- Full Genomes results by Ricardo Costa de Oliveira J1b M365+ blog, 22 November 2013.
Blog posts
- Kane J NGS Y Chromosome Alignment on GRCh38. Irish Type 2 - Kane blog, 22 November 2015.
Privacy policy, terms and conditions
This information is not yet available on the homepage and was given by Justin Loe in the Facebook page in December 2013.
- DNA: sent to the lab
- Data: data is stored where analyses are performed and in our site
- Data Access: not published, via website or direct delivery
- IDs: labs only receive anonymous ids, not names
External links
- Full Genomes official website
- Full Genomes demo tree
- Full Genomes Facebook group
- Full Genomes on Google+
- Fully Y-chromosome sequencing Phase III pilot A discussion on the Anthrogenica Forum about the Full Genomes test
- Manuelcorpas.com
References
- ↑ Company Overview of Full Genomes Corporation, Inc.: http://www.bloomberg.com/research/stocks/private/people.asp?privcapId=246878958
- ↑ Loe J. Personal communication to Debbie Kennett, 6 August 2017.
- ↑ Loe J. Personal communication to Debbie Kennett. 25 November 2017.
- ↑ Justin Allen Loe. Message posted 2013-11-09 in Facebook group, https://www.facebook.com/groups/fullgenomesY/
- ↑ Greg Magoon comparison table, published in FGC Facebook group, http://i.imgur.com/Gq7jDrX.png
- ↑ ADAMOV, D., GURYANOV, V., KARZHAVIN, S., TAGANKIN, V., URASIN, V.. Defining a New Rate Constant for Y-Chromosome SNPs based on Full Sequencing Data. The Russian Journal of Genetic Genealogy (Русская версия), mar. 2015. Available at: http://rjgg.molgen.org/index.php/RJGGRE/article/view/151/175. Date accessed: 06 May. 2015.
- ↑ Feedback from a whole genome customer. June 2015. http://www.anthrogenica.com/showthread.php?742-Full-Y-Chromosome-Sequencing-Phase-III-Pilot&p=89764&viewfull=1#post89764
- ↑ Sample FGC whole genome: (Y chromosome data from whole genome), July 2015. http://www.anthrogenica.com/showthread.php?742-Full-Y-Chromosome-Sequencing-Phase-III-Pilot&p=94263&viewfull=1#post94263
- ↑ Greg Magoon about prototype results: "102.8 million variant sites with read depth ≥8, 106.5 million with at least one mapped read.", E-Mail Mar 23, 2015
- ↑ McConnell et al 2013: Mosaic copy number variation in human neurons. http://www.ncbi.nlm.nih.gov/pubmed/24179226
- ↑ Justin Loe, anthrogenica Forum, 2016-03-20, http://www.anthrogenica.com/showthread.php?742-Full-Y-Chromosome-Sequencing-Phase-III-Pilot&p=146746#post146746