Mitochondrial DNA tests
From ISOGG Wiki
A mitochondrial DNA test (mtDNA test) traces a person's matrilineal or mother-line ancestry using the DNA in his or her mitochondria. mtDNA is passed down by the mother unchanged, to all her children, both male and female. A mitochondrial DNA test, can therefore be taken by both men and women. If a perfect match is found to another person's mtDNA test results, one may find a common ancestor in the other relative's (matrilineal) "information table".
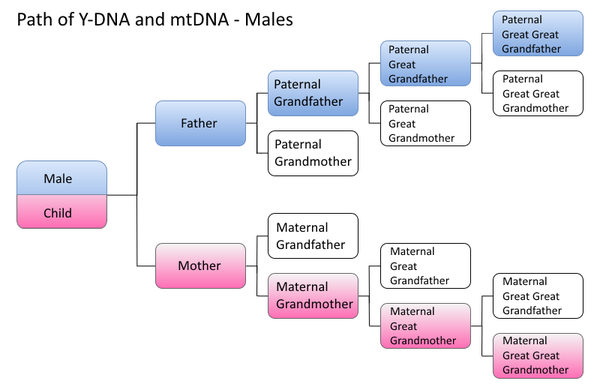
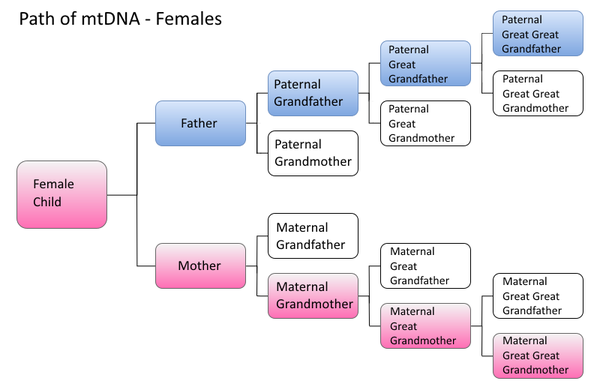
Path of mtDNA transmission
Males inherit mtDNA (shown in pink) from their mother but do not pass it on to their children. Males inherit Y-DNA (shown in blue) from their father. They pass on Y-DNA to their sons but not their daughters.
Females inherit mtDNA (shown in pink) from their mother. They pass on mtDNA to both their male and female children. Females do not inherit Y-DNA (shown in blue) from their father.
See also the animation on mitochondrial DNA on the Learn Genetics website which provides a good explanation of the inheritance of mtDNA.
What gets tested
mtDNA by current conventions is divided into three regions. They are the coding region (00577-16023) and two hyper-variable regions (HVR1 [16024-16569], and HVR2 [00001-00576]). All test results are compared to the mtDNA of a European in haplogroup H2a2a. This early sample is known as the Cambridge Reference Sequence (CRS). An updated reference sequence was subsequently published and samples are now compared to the revised Cambridge Reference Sequence (rCRS). A list of single-nucleotide polymorphisms (SNPs) is returned. The relatively few "mutations" or "transitions" that are found are then reported simply as differences from the CRS, such as in the examples just below.
In 2012 Behar et al proposed using an inferred ancestral reference sequence known as the Reconstructed Sapiens Reference Sequence.[1] Bandelt et al (2013) questioned the use of the RSRS and proposed the retention of the rCRS.[2] Genetic genealogy companies have continued to report results in comparison with the rCRS. Family Tree DNA report results in comparison with both the rCRS and the RSRS. The Genographic Project currently reports results in comparison with the RSRS.
The two most common mtDNA tests are a sequence of HVR1 and a sequence of both HVR1 and HVR2. Some mtDNA tests may only analyze a partial range in these regions. With falling prices the preferred option is now to test both HVR1 and HVR2 as a minimum. Family Tree DNA stopped offering the basic HVR1 test in May 2013.
Many people are now choosing to have the full mitochondrial genome sequenced (scientifically called the mtDNA genome or mitogenome). This is at times called the mtDNAFullSequence, the Mega Test, or the full mitochondrial sequence (FMS) test. An FMS test will report results for all 16,569 bases in the mtGenome and will provide the most detailed subclade assignment. If two people have an exact FMS match they will generally share a common ancestor within the last 22 generations (about 550 years).[3] Conversely it is sometimes possible for mothers and daughters or siblings to have differences in their mtDNA. These differences usually take the form of a heteroplasmy.
Family Tree DNA and GeneBase are the only two companies that currently offer the FMS test. A full sequence test was previously available from the now defunct company Argus Biosciences.
Some companies and organisations, such as the Genographic Project, 23andMe and Living DNA, use microarray chips which provide a scan of selected SNPs from across the entire mtDNA genome. The SNPs are chosen for the phylogenetic relevance and can provide a fairly detailed haplogroup assignment. However the results from these tests are not compatible with the genealogical matching databases.
Understanding test results
The most basic of mtDNA tests will sequence hyper-variable Region 1 (HVR1). HVR1 nucleotides are numbered 16024-16569. Some test reports might omit the 16 prefix from HVR1 results, i.e. 519C and not 16519C.
| Region | HVR1 | HVR2 |
|---|---|---|
| Differences from rCRS | 111T,223T,259T,290T,319A,362C | Not Tested |
More extensive tests will also sequence Hyper Variable Region 2 (HVR2). HVR2 nucleotides are numbered 00001-00576.
| Region | HVR1 | HVR2 |
|---|---|---|
| Differences from rCRS | 111T,223T,259T,290T,319A,362C | 073G,146C,153G |
Family Tree DNA customers who have ordered a full mitochondrial sequence test from FTDNA can order a custom mtDNA report from ISOGG member Dr Ann Turner to help them understand their results.
mtDNA tree
Phylotree provides a public copy of the mtDNA tree based on samples uploaded to the public GenBank database.
Family Tree DNA provides a public mtDNA haplotree that follows the Phylotree nomenclature and draws from over 170,000 mitochondrial full sequences tested at FTDNA.
Phylotree is now on Build 17. Updates are released every couple of years. Genetic genealogists who have taken mtDNA tests with different companies will sometimes find that they receive different haplogroup assignments. This is because the companies are using different versions of the mtDNA tree. Family Tree DNA updated to Build 17 towards the end of March 2017. 23andMe upgraded their haplogroup reports in June 2017.
A large and detailed mtDNA phylogeny can be seen on the Mitomap website.
An up-to-date haplogroup assignment can be obtained from James Lick's mtHap utility. This tool is usually updated within 24 hours of the release of each new Phylotree build.
Haplofind is an alternative and less sophisticated tool more suited for batch processing mtDNA full sequence results. It does provide a list of mutations with reported and associated medical implications.
Understanding mtDNA testing
- Understanding mitochondrial DNA testing by Steve Handy. DNA Genealogical Experiences and Tutorials blog, 4 November 2012.
- DNA testing for genealogy - getting started Part Two: mitochondrial DNA by CeCe Moore, Geni.com blog, 25 July 2012
mtDNA databases
- Ancient mtDNA database
- GenBank - for FMS results only
- Charles Kerchner's mtDNA test results log
- Ian Logan's Website of mtDNA Sequences Submitted to GenBank
FTDNA resources
- DNA test basics: maternal lineages
- My FTDNA mtDNA matches page
- My FTDNA user guide: mtDNA pages in the FTDNA Learning Center
- mtDNA pages in the FTDNA Learning Center
- Scientific collaboration mtDNA full sequence results donation
- FTDNA's list of mtDNA haplogroup-defining mutations
Blog posts
- Mitochondrial DNA - a blast from the past by Robin Wirthlin, The Family Locket, 1 February 2021.
- How to make matches with mitochondrial DNA by Linda Jonas, The Ultimate Family Historian, 8 August 2020. The article includes detailed instructions on how to get your mtDNA results into all the available databases as well as some interesting insights into the history of mtDNA testing.
- Mitochondrial DNA: Part 1 – Overview by Roberta Estes, first of a five-part series, DNAeXplained, 16 May 2019.
- Demystifying DNA 3: mtDNA testing by Karen Cummings, Genealogical Ponderings, 2 August 2018.
- Heteroplasmies and Poly-Cytosine Stretches – An mtDNA Case Study by Blaine Bettinger, The Genetic Genealogist, 21 April 2018.
- DNA basics: mitochondrial DNA by Leah Larkin, The DNA Geek, 19 April 2018.
- Using mtDNA for genealogy - a case study by Leanne Cooper, Leanne Cooper Genealogy, January 2018.
- Mitochondrial DNA: Connecting Generations. LegacyTree Genealogists blog, 28 December 2016.
- Mitochondrial - the maligned DNA by Roberta Estes, DNAeXplained, 29 March 2014.
- Your privacy and showing your mtDNA coding region results by Rebekah Canada, Haplogroup blog, 9 July 2013.
- Mitochondrial DNA testing at a new low price by Debbie Kennett, Cruwys News blog, 10 May 2013 (article includes a discussion of the merits of the different types of mtDNA test)
Articles
- Parker Wayne D. Using mitochondrial DNA for Genealogy. US National Genealogical Society NGS Magazine 39 (October-December 2013): 26-30.
- Smolenyak M. The ABCs of mtDNA
- Roderick TH, King M-C, Anderson RC (1992). Mitochondrial DNA: A genetic and genealogical study. NEXUS, October-November issue, New England Historic Genealogical Society, 22 November 1992.[Footnote 1]
Scientific papers
- Wei, Tuna et al. (2019) Germline selection shapes human mitochondrial DNA diversity. Science 364(6442): eaau6520.
- Andersen & Balding (2018) How many individuals share a mitochondrial genome? PLoS Genetics 14(11): e1007774.
- Toomas Kivisild (2015) Maternal ancestry and population history from whole mitochondrial genomes. Investigative Genetics 2015(6): 3.
- King TE et al. (2014) The identification of Richard III. Nature Communications 5; 5631. mtDNA testing combined with genealogical research played a significant part in the identification.
- Ma, Coarfa et al. (2014) mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genomics 2014(15): 257.
- Behar, van Oven et al. (2012) A “Copernican” Reassessment of the Human Mitochondrial DNA Tree from its Root. American Journal of Human Genetics 90(5): 936.
- Balloux F (2009). The worm in the fruit of the mitochondrial DNA tree. Heredity 104: 419-420.
- Henn, Gignoux et al. (2009) Characterizing the Time Dependency of Human Mitochondrial DNA Mutation Rate Estimates. Molecular Biology and Evolution 26(1): 217-230.
- Loogväli, Kivisild et al. (2009) Explaining the Imperfection of the Molecular Clock of Hominid Mitochondria. PLoS ONE 4(12): e8260.
- Robert Carter (2007) Mitochondrial diversity within modern human populations. Oxford Academic, Nucleic Acids Research 35(9): 3039–3045.
- Ballard JWO, Whitlock MC (2004). The incomplete natural history of mitochondria. Molecular Ecology 13: 729–744.
Resources
- The mtDNA encyclopedia of human origins
- Ian Logan's mtDNA website
- mtDNA tutorial from Genebase
- Mitomap A compendium of polymorphisms and mutations of the human mitochondrial DNA
- mtDNA resources from SMGF (website preserved in the Wayback Machine)
- Mitochondrial sequencing Information from Illumina
- SNPedia—mtDNA Haplogroup Includes associated genosets and a table showing major haplogroup defining variants with links to explore variants for subclades
- Wikipedia article on mitochondrial DNA
- Wikipedia article on human mitochondrial genetics
Videos
- Mitochondrial DNA A video from the Sorenson Molecular Genealogy Foundation now hosted on the Learn Genetics website
A video of a presentation by ISOGG member Maurice Gleeson showing how mitochondrial DNA testing can be used in family history research:
Bill Hurst, the administrator of the mtDNA haplogroup K, project, describes how he successfully used mtDNA testing to unravel the tale of two sisters:
See also
- ISOGG Wiki mtDNA Portal
- Before You Buy
- Matriline
- mtDNA testing comparison chart
- mtDNA haplogroup projects
- mtDNA tools
- Custom mitochondrial DNA reports
- Mutation rates
- BritainsDNA haplogroup nicknames
- Oxford Ancestors haplogroup nicknames
- Success stories
References
- ↑ Behar DM, Van Oven M, Rosset S et al. A "Copernican" reassessment of the human mitochondrial DNA tree from its root. American Journal of Human Genetics 2012; 90 (5): 936.
- ↑ The case for the continuing use of the revised Cambridge Reference Sequence (rCRS) and the standardization of notation in human mitochondrial DNA studies Bandelt HJ, Kloss-Brandstatter A, Richards MB et al. Journal of Human Genetics 2014 59(2): 66-77.
- ↑ How do I tell how closely I am related to a mitochondrial DNA match? Family Tree DNA Learning Center. Accessed 17 April 2106.
Footnotes
- ↑ The New England Historic Genealogical Society published NEXUS until the Nov-Dec.1999 issue. They replaced it with New England Ancestors which in turn was succeeded by the winter 2010 issue of American Ancestors.
Licence
This article is licensed under the GNU Free Documentation Licence. It incorporates material from the Wikipedia article "Genealogical DNA test".